PhD project
Project 1 Nudivirus replication mechanism (NL)
The research project "Nudivirus replication mechanisms" aims to unravel the replication mechanisms and gene functions of nudiviruses (Nudiviridae). These invertebrate-infecting viruses are phylogenetically related to endogenous bracoviruses (Polydnaviridae) found in parasitoid wasps. The Heliothis zea nudivirus 1 (HzNV-1) genome was found in its host as a persistent infection. Experiments with nudiviruses and bracoviruses aim to reveal whether these viruses have similar replication processes.
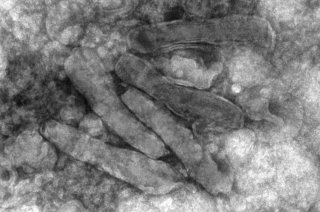
One of the most diverse virus families found in insects are nudiviruses (from Latin, nudi = naked or uncovered) of the family Nudiviridae. They have been found to infect different insect orders, including crickets, beetles, moths and flies, but also marine invertebrates, such as lobsters and shrimp. Nudivirus infections in invertebrates can reduce their hosts' fitness and under certain conditions lead to their death. Therefore, it is important to study the impact of these diverse viruses to prevent devastating losses in aquacultures and the uprising industry of insect rearing. Conversely, the harmfulness of a beetle-infecting nudivirus (Oryctes rhinoceros nudivirus, OrNV) has been used to control infestations of this insect pest in palm tree cultures in parts of Oceania, providing another relevant reason to study this virus family.
The species Heliothis zea nudivirus comprises two strains, HzNV-1 and HzNV-2. Its name originates from its natural host Helicoverpa (formerly Heliothis) zea, commonly known as the corn earworm. Caterpillars of this moth pose a threat to corn and other crop plants. HzNV-2 is sexually transmitted between individuals or to the offspring via infected ovaries, and causes disease in larvae and adults of H. zea. HzNV-1, on the other hand, was isolated as a persistent viral infection in a cell line derived from H. zea ovarian tissue and was not capable of infecting actual insects. HzNV-1 can induce latent and lytic infections in different insect cell lines and may persist in the host cells through viral genome integration.
It was shown that the integration of an ancestral nudivirus into the genome of an ancestor braconid wasp 100 million years ago gave rise to the clade of endogenous bracoviruses. Therefore, it is reasonably assumed that the replication mechanisms and machinery of bracoviruses derive from its nudiviral ancestor. However, little is known how bracovirus replication is maintained after viral genome integration and what genes facilitate crucial functions in this state. Our main research aims to reveal similarities in the gene functions of nudiviral and bracoviral replication by conducting comparative studies with the latest molecular, genomic analytical tools and microscopic techniques.